MATERIALS
REAGENTS (Note 1)
DNA extraction kit (Qiagen DNeasy Blood & Tissue kit, cat. 69504/69506)
GoTaq® G2 DNA Polymerase (Promega, cat. M7845)
Nuclease-free water (Qiagen, cat. 129114)
dNTP mix (Promega, cat. U1511)
Primers (see Table 1, ordered at Integrated DNA Technologies)
LightCycler® 480 Probes Master, 2x concentrated (Roche Applied Science, cat. 04707494001)
EQUIPMENT
Thermal Cycler Applied Biosystems® 2720 (cat. 4359659)
LightCycler® 480 System II (Roche, cat. 05015278001)
Primers |
Sequence |
1st PCR (Alu-Gag PCR) |
|
Forward primer Alu(1) |
5’-GCCTCCCAAAGTGCTGGGATTACAG-3’ |
Reverse primer Gag(1) |
5’-GTTCCTGCTATGTCACTTCC-3’ |
2nd PCR (qPCR) |
|
Forward primer RU5 (R Forward)(2) |
5’-TTAAGCCTCAATAAAGCTTGCC-3’ |
Reverse primer RU5 (U5 Reverse)(2) |
5’-GTTCGGGCGCCACTGCTAGA-3’ |
Probe(3) |
5'-CCAGAGTCACACAACAGACGGGCACA-3' |
PROCEDURE (download as PDF)
Isolate genomic DNA from the target cells using the DNA extraction kit. To estimate the volume of material used for the input in PCR reaction, one can either use a fixed number of input cell equivalents, e.g. 7500 equivalents per reaction as described by (1) Liszewski et al. 2009, , or adjust input to the amount of total HIV DNA, as estimated by a prior total HIV DNA quantification by real time or digital PCR (Note 2).
INTRODUCTORY INFORMATION
The Alu-HIV PCR is designed to quantify integrated proviral HIV DNA through a nested PCR setup of two consecutive PCR amplification rounds. In the first PCR forward primers are used for the human Alu repeat sequence in combination with a reverse primer, targeting an HIV specific sequence (fig. 1). This reaction only leads to a logarithmic amplification of HIV DNA when this DNA is integrated, and thus in the vicinity of an Alu sequence. Unintegrated linear or episomal HIV DNA is amplified in a linear faschion. The second PCR is composed of a quantitative PCR (qPCR) that amplifies an HIV specific sequence which is nested within the amplicon of the first PCR. To enable the differentiation of the background signal originating from unintegrated HIV DNA, a control is run in parallel using only an HIV reverse primer to mimic the kinetic PCR that would take place when unintegrated HIV DNA is present.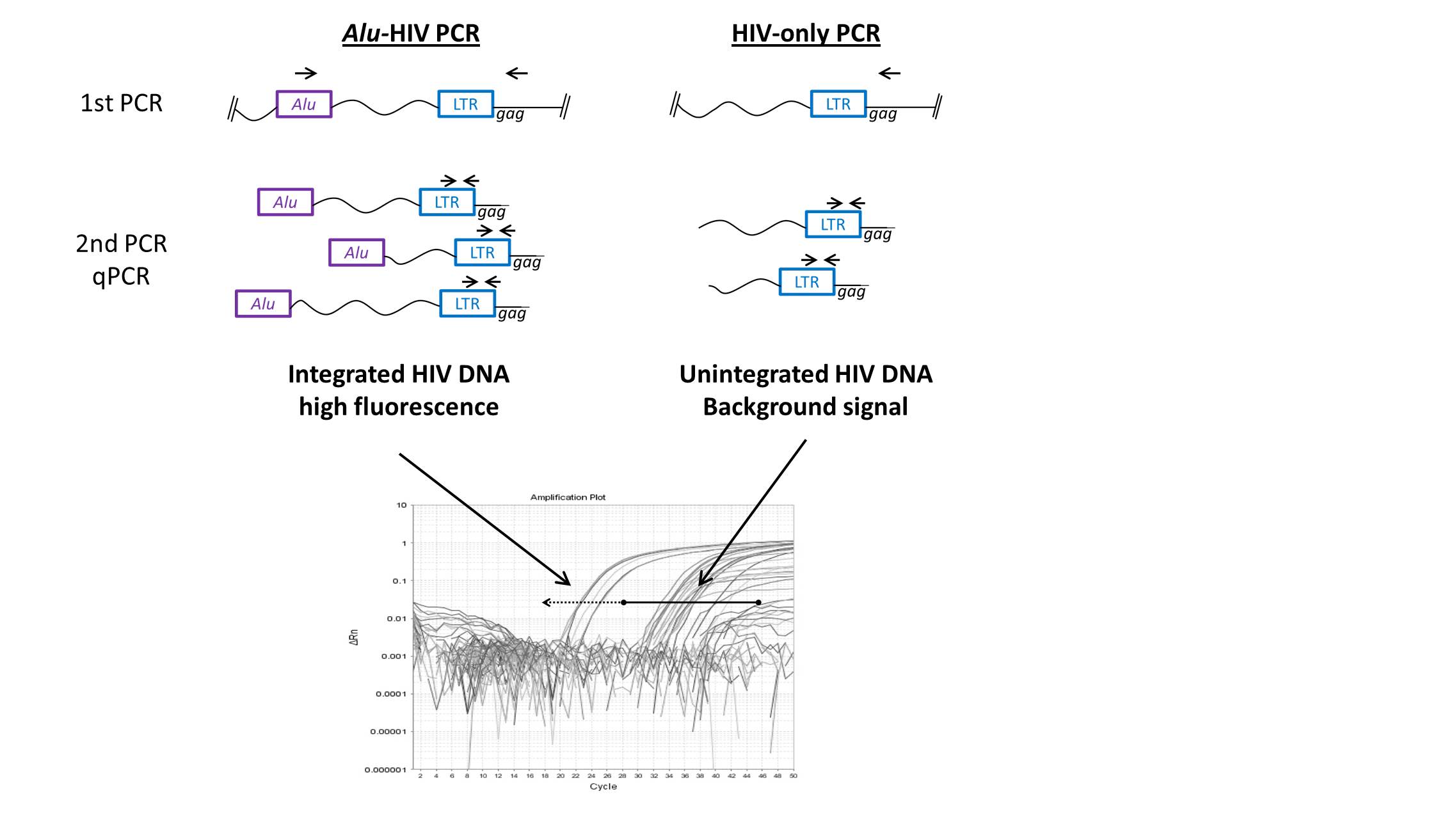
Depending on the origin of the DNA for analysis (patient samples or cell cultures), the input for the 1st PCR need to be selected. For patient samples, the reaction works best if the input is in the range of 1-2 HIV DNA copies/well (Note 3).
To maximize the power of the analysis, 40 replicates for the Alu-Gag primers combination and minimally 20 for Gag only primer (background) are performed. If enough material is present, an equal number of Gag-only and Alu-Gag replicates is preferred (Note 4).
Prepare the mix for the appropriate number of replicates consisting of:
PROMEGA GoTaq |
Alu-gag mix |
gag-only mix |
Buffer (5X) |
4 | 4 |
dNTP's (10mM) |
0.4 | 0.4 |
H2O |
4 | 4.2 |
GoTaq |
0.2 | 0.2 |
Forward primer (nM) |
0.2 | |
Reverse primer (nM) |
1.2 | 1.2 |
Distribute the mix in a 96 well plate (10 µl per well) taking the amount of replicates of both Alu-Gag and Gag only into account, add 2 wells of each for the positive and negative controls.
Add 10 µl of the DNA solution to each well except for the controls. In the controls, add 9 µl of water and 1 µl of control DNA (from the integration standard) to the positive controls, and 10 µl of water to the negative controls.
Seal the plate and run the PCR protocol in the Thermal Cycler, using the following cycling conditions:
Reaction details |
Step |
°C | time |
| 1X | initial denaturation | 95 | 02:00 |
| denaturation | 95 | 00:15 | |
| 40X | annealing | 50 | 00:15 |
| elongation | 70 | 03:30 |
THE SECOND PCR REACTION
The relative quantification is performed with a qPCR on the PCR product from the first PCR step
Prepare the qPCR mix consisting of:
|
µl/reaction |
PCR mastermix |
5 |
H2 |
2 |
Probe (10 µM) |
0.2 |
Forward primer (10 µM) |
0.4 |
reverse primer (10 µM) |
0.4 |
Distribute the mix in a 384 well plate, 8 µl per well.
To each of the wells, add 2 µl of the 1st PCR solution from a corresponding well. Maintain the number of replicates, use each replicate only once.
Run the reaction on LightCycler® 480 System II (Roche).
The cycling conditions are:
Reaction details |
Step |
°C | time |
| 1X | initial denaturation | 95 | 05:00 |
| 45X | denaturation | 95 | 00:15 |
| annealing | 60 | 01:00 |
ANALYSIS
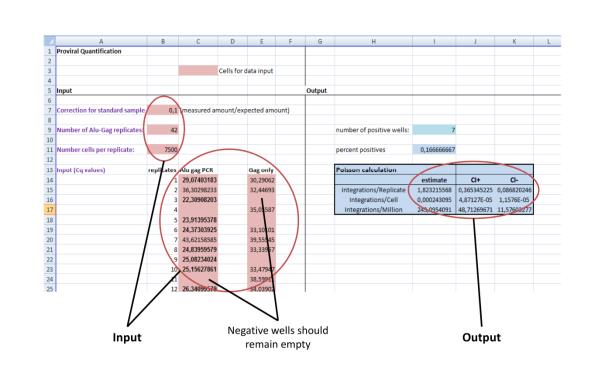
To assess the number of integrations per cell, the excel template (Fig. 2) can be used. The raw quantitative output of the Alu-HIV PCR as Cq values (Cycle of quantification, also termed Ct) should be inserted as described below.
REFERENCES
De Spiegelaere W, Malatinkova E, Lynch L, Van Nieuwerburgh F, Messiaen P, O'Doherty U, Vandekerckhove L: Quantification of Integrated HIV DNA by Repetitive-Sampling Alu-HIV PCR on the Basis of Poisson Statistics. Clin Chem 2014. url
Yu JJ, Wu TL, Liszewski MK, Dai J, Swiggard WJ, Baytop C, Frank I, Levine BL, Yang W, Theodosopoulos T, O'Doherty U: A more precise HIV integration assay designed to detect small differences finds lower levels of integrated DNA in HAART treated patients. Virology 2008;379:78-86. url
Yun Z, Frederiksson E,Sönnerborg A: Quantification of human immunodeficiency virus type 1 proviral DNA by the TaqMan real-time PCR assay. Journal of Clinical Microbiology 2002;40:3883-3884. url
NOTES
These reagents and materials are being used by us. An alternative method for Alu-HIV PCR has also been described by Liszewsky et al (2009)
Using a fixed number of cell equivalents provides a standardized reaction. However, in patient samples with limited HIV DNA, most reactions become negative which may limit the quantitative power of the Poisson calculation afterwards. By estimating the number of HIV DNA templates before Alu-HIV PCR, the concentration of HIV DNA templates can be estimated to get an optimal Poisson quantification.
We generally start with 10 DNA copies for cell cultures per reaction, considering the fact that cell culture generally have a higher concentration of unintegrated HIV DNA, compared to patient derived samples.
We use a cut-off of five positive Gag-only PCR reactions to estimate the background. If this number is not reached, the amount of Gag-only PCR replicates should be augmented.